Current versions of IGB supported: 8.3 and 8.4. If using version 8.3, check in the plugin description (plugin tab) which version to load (the selection is automatic starting from version 8.4).
The official page for the MI bundle will soon move to its github repository: http://arnaudceol.github.io/igb-mi-bundle/
From genome to molecular interactions, a plug-in to map next generation sequencing data to molecular interactions and their structures with the Integrated Genome Browser.
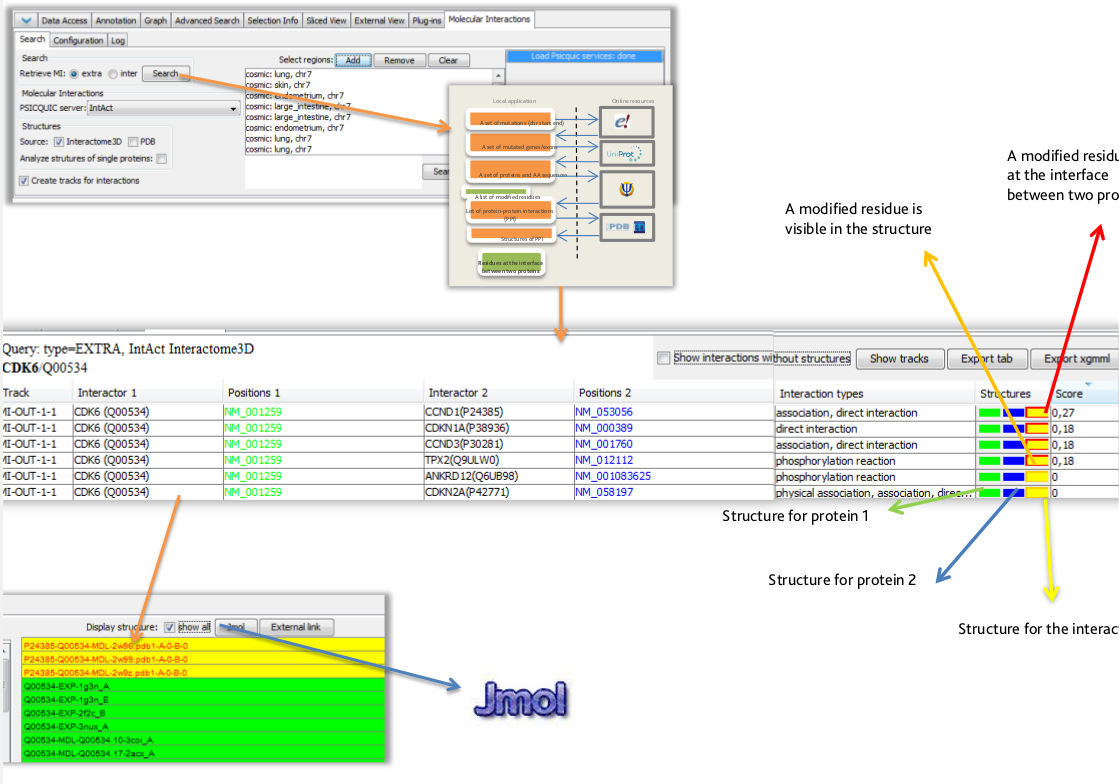
Next-generation sequencing has been applied to identify genomic variations possibly associated to many diseases. The next challenge consists in identifying the role of those mutations. Mutations located in protein interaction interfaces are for instance often associated with loss-of-function or gain-of-function. The identification of such residues requires the integration of disseminated biological data and bioinformatics tool. Fortunately, the possibility to extend the existing genome browsers allows us to integrate complex frameworks and make them available through already widely adopted tools.
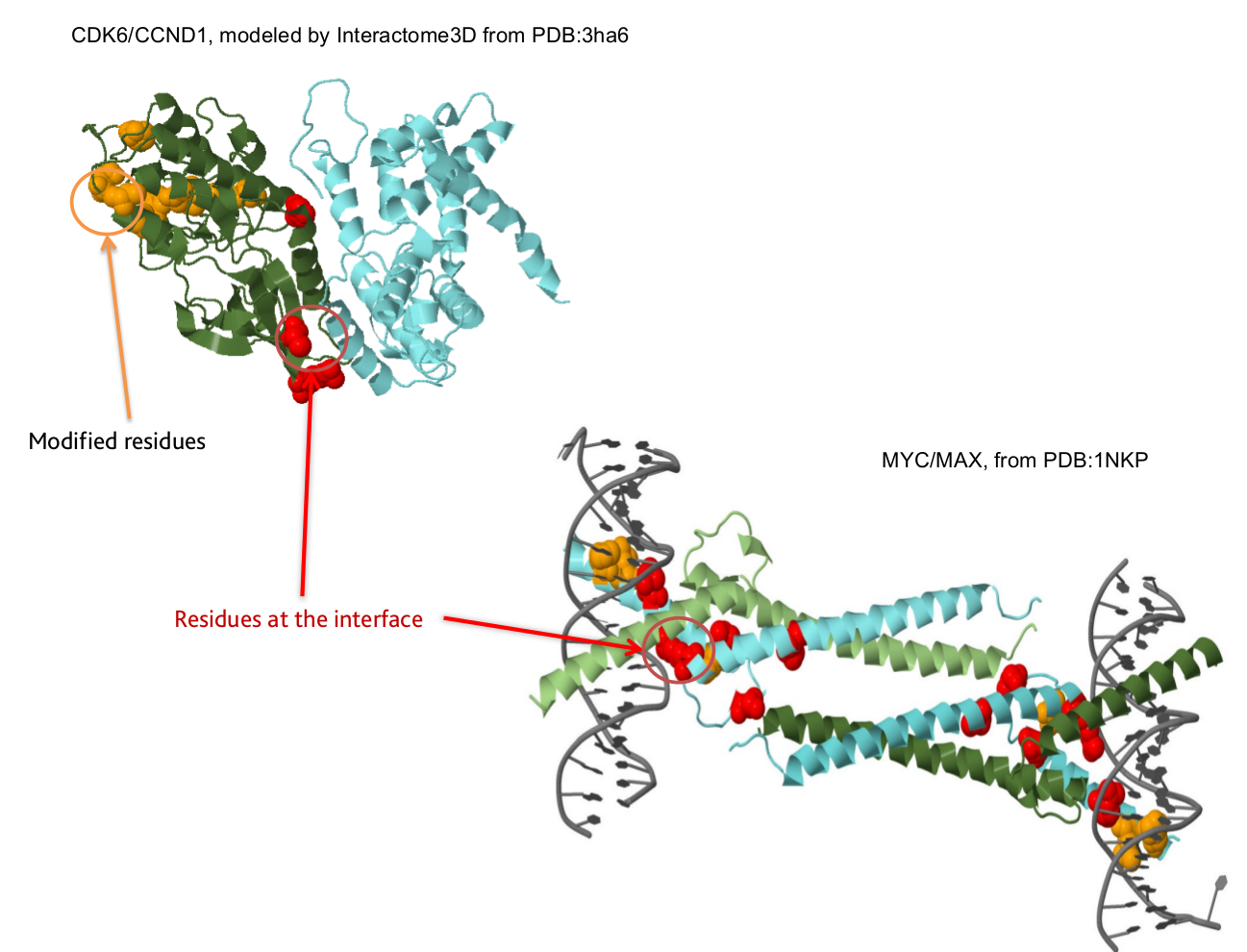
We have developed an extension for the Integrated Genome Browser, a widely used tool for the visualization and analyzes of genomic data, which maps and visualizes genomic regions in molecular interactions structures (protein-protein, protein-DNA, protein-RNA and protein-small molecule interactions) and allows the researcher to make educated guesses about the functional impact of somatic mutations.