THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE, TITLE AND NON-INFRINGEMENT. IN NO EVENT SHALL THE COPYRIGHT HOLDERS OR ANYONE DISTRIBUTING THE SOFTWARE BE LIABLE FOR ANY DAMAGES OR OTHER LIABILITY, WHETHER IN CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
This tutorial will guide you through the usage of the MI Bundle, from its installation to the display of cancer mutations in a protein interaction structure.
Install and run IGB.
Go to http://bioviz.org/igb/ and click Download now to download IGB. You need Java to be installed in order to run IGB (you can find instruction on how to do it at https://www.java.com/en/download/index.jsp). In IGB, go the the plugin tab (1), and press the repositories... button (2).
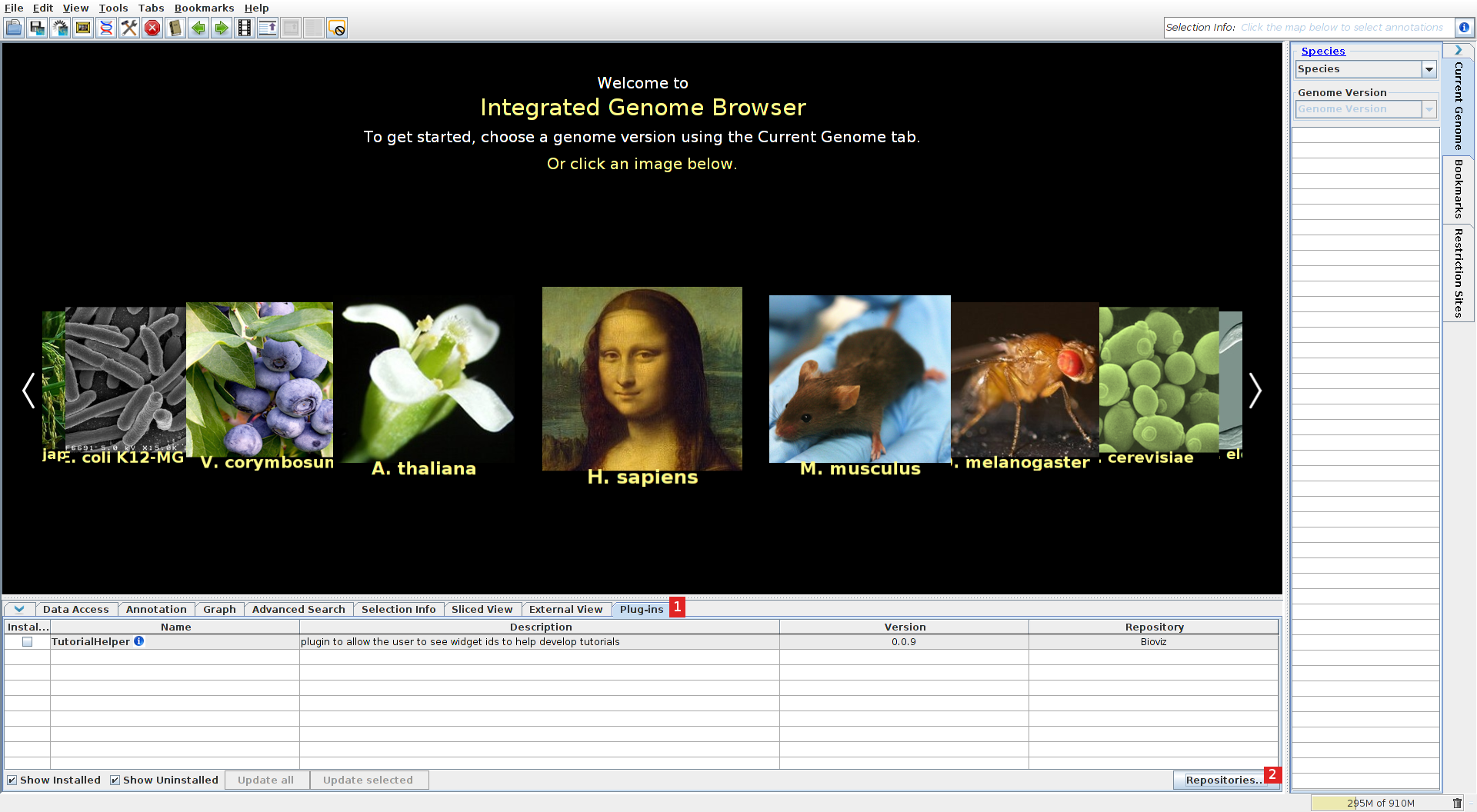
Add the IIT plugin repository
Add (1) the repository "http://cru.genomics.iit.it/igb/plugins/" (2).
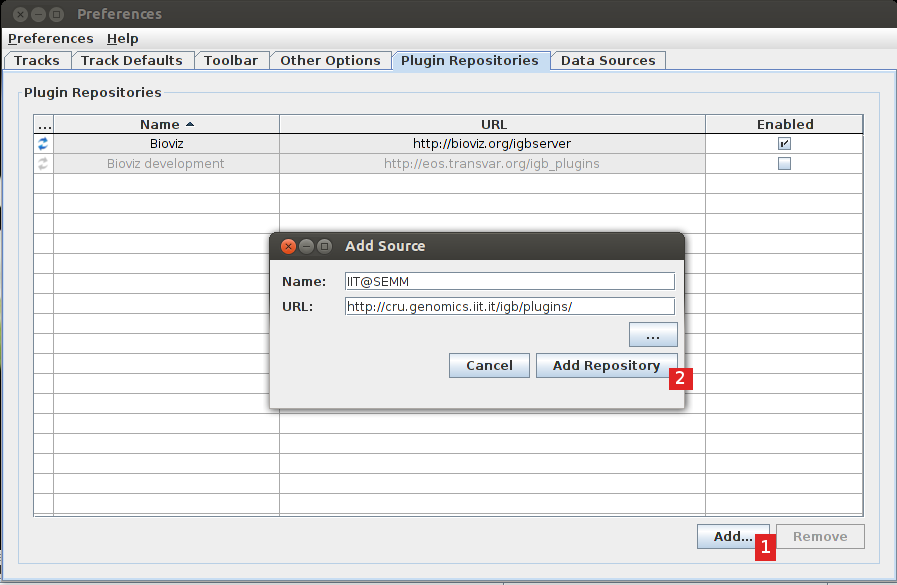
The plugin should now appear in the list, select the check box to install it (1).
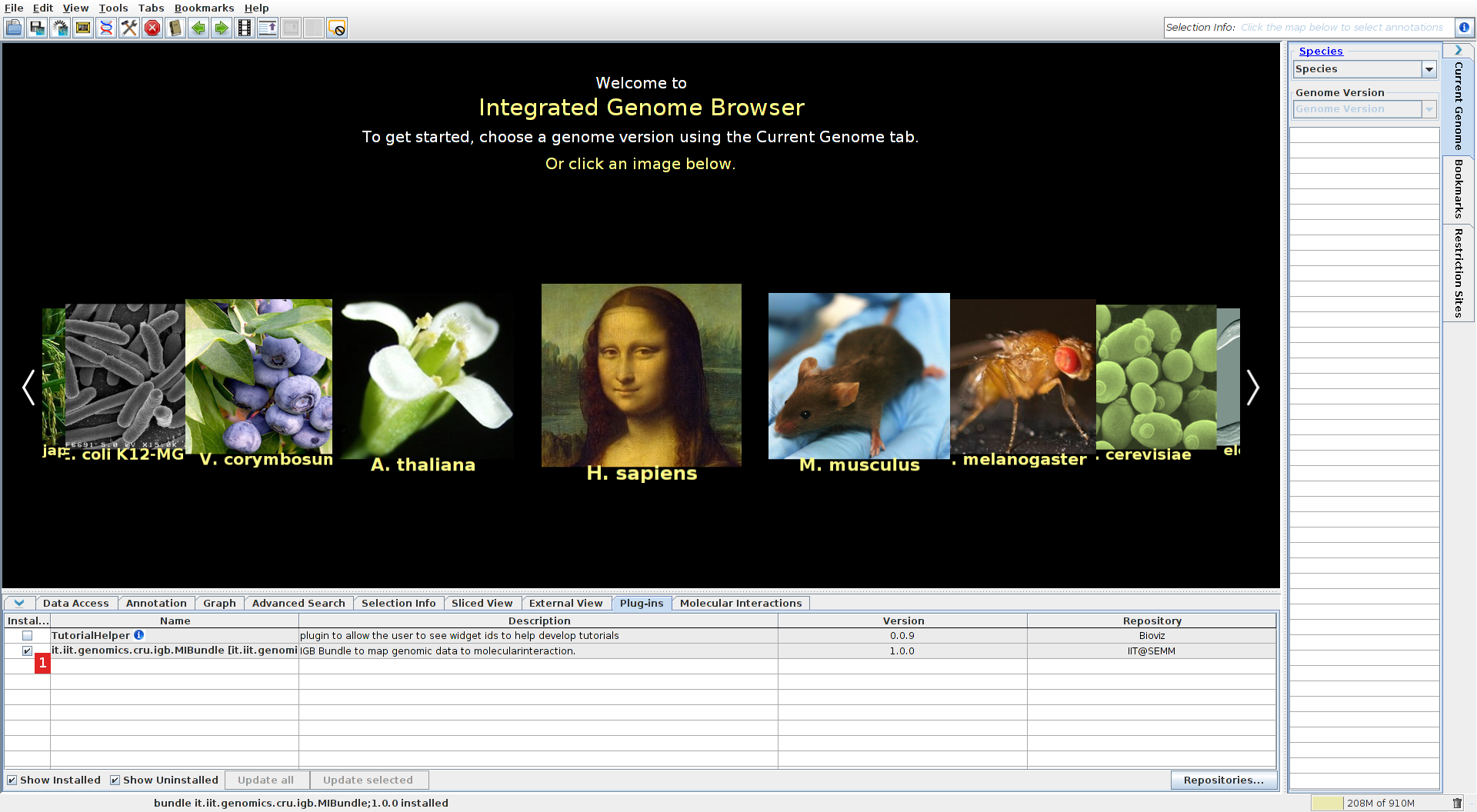
Select a species
Select the human genome assembly (select the "current genome" menu on the right, then species: Homo sapiens and the last version, or directly click on the Mona Lisa portrait). Go to the data access tab and ensure that refGene or ensGene is selected (1) and that the load mode is set to genome (2) (this is necessary for the MI Bundle to identify the genes for interacting proteins).
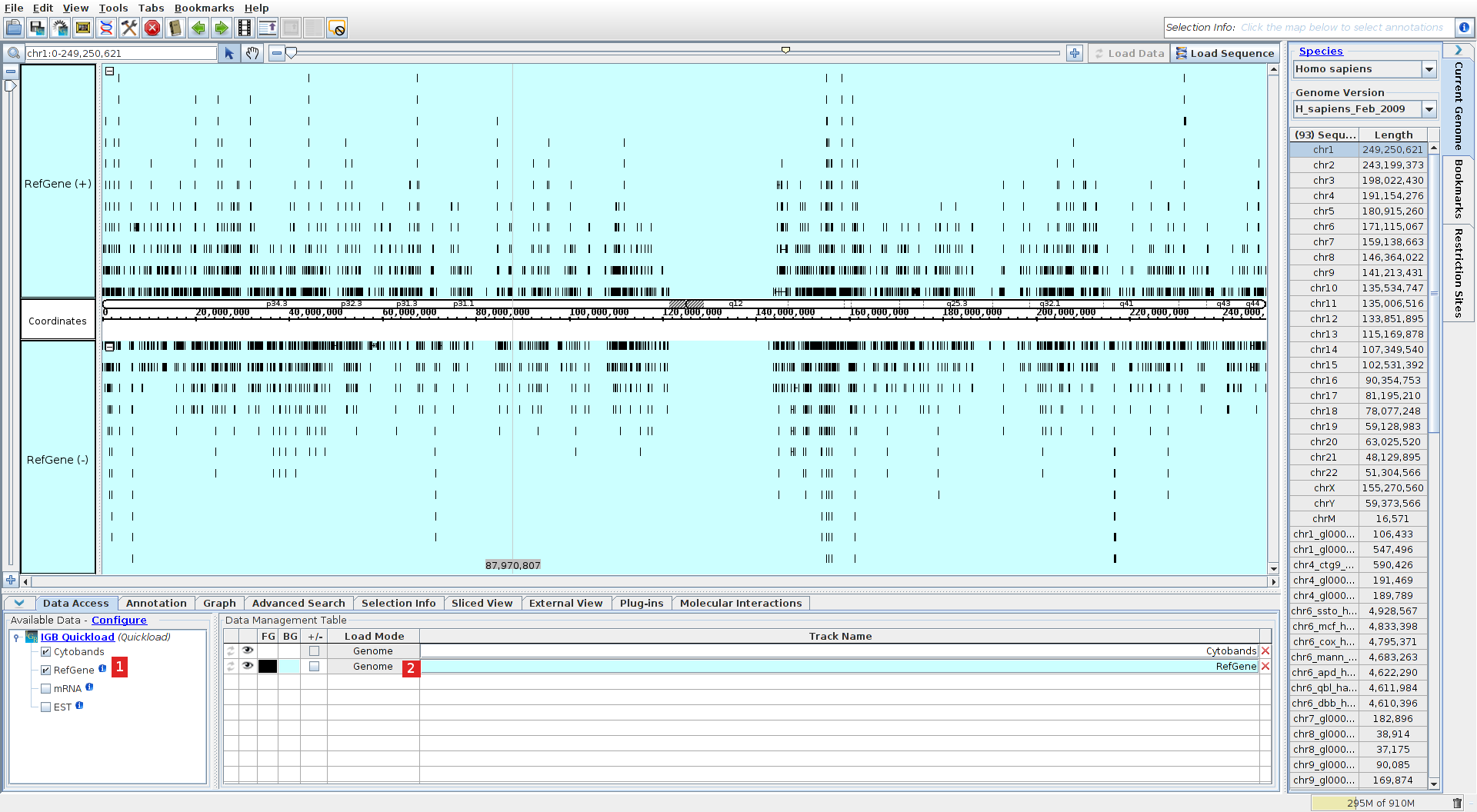
Load variations from an external file
Load mutations from Clinvar (a repository for sequence variation and its relationship to human health) in vcf format: go to File, open URL... and fill with the URL of the VCF file: ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/clinvar.vcf.gz.
Search a gene
Go to advance search (1) and enter RUNX1, double click on the RUNX1 gene entry (2). Press the load data button inthe top-right corner of the browser. All mutations in the gene's region should now be loaded.

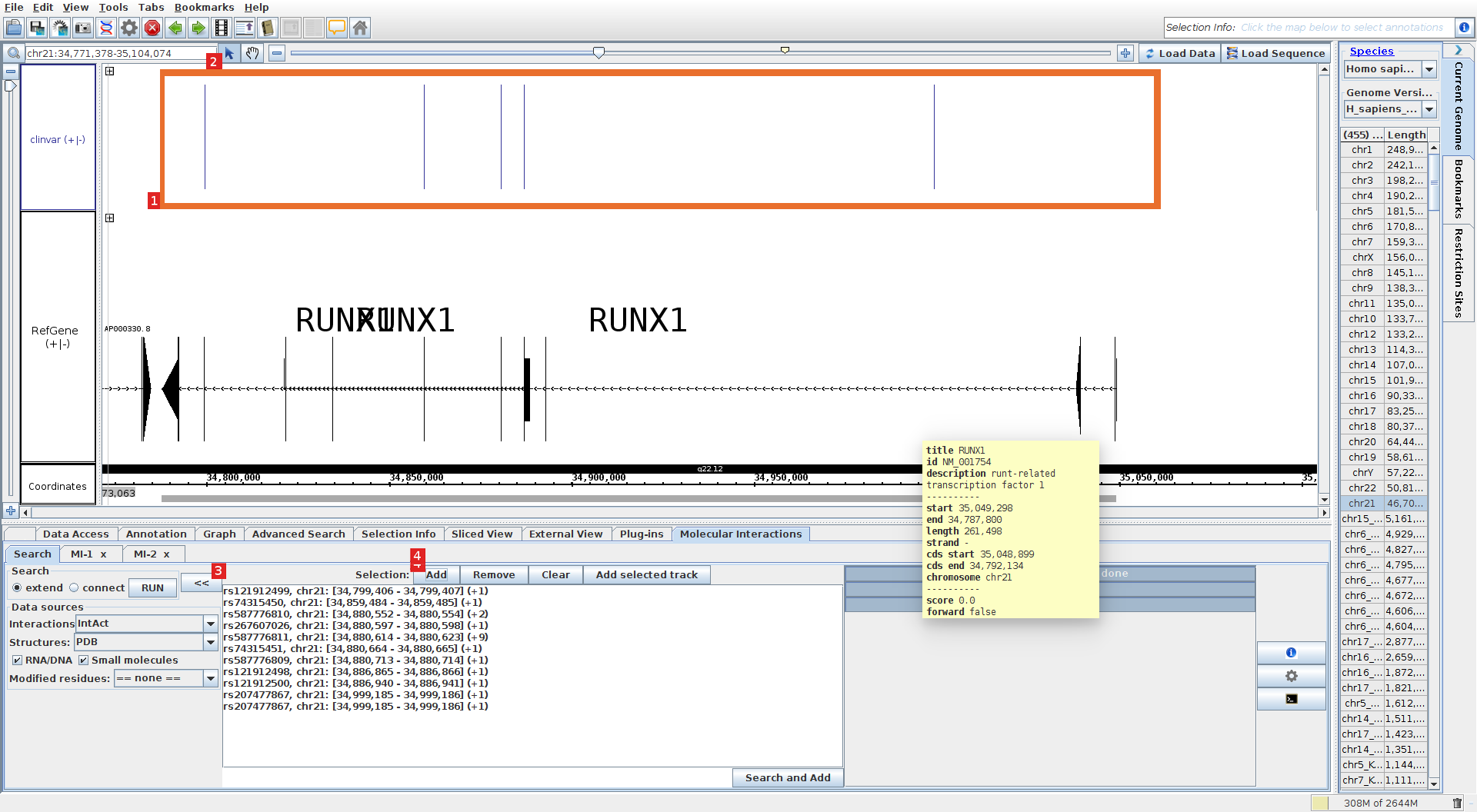
Select mutations and run the analyzes.
Select all mutations (1) (if the hand is selected, you hay have to select the arrow in the top-left corner of the browser, beside the chromosome region field (2)). Go to the Molecular Interaction tab, press >> to show the search box and press add (3), the mutations will be added to the search box. Check the small molecules and DNA/RNA check boxes to include those types of molecules. Press the RUN button (4) to run the analyzes. (in alternative you may press directly the RUN button after selecting the regions with the mouse without displaying first the search box).
Results, display structure.
The results are displayed in a new table. Click on the row that contains the interaction between RUNX1 and DNA, the list of available structures will be filled.
Press the Jmol button to display the first structure (1). Right click on the Jmol window to access all Jmol options (2).
